Introduction
Acute Lymphoblastic Leukemia (ALL) is the most common malignancy in children and represents 75-80% of leukemia cases. Despite its recognized role in the initiation of the disease, the bone marrow (BM) microenvironment is not well understood especially in its non-cellular component.
High-throughput protein detection represent an opportunity to unveil new biomarkers for leukemia diagnosis, prognosis, monitoring, and to potentially identify biological targets.
Methods
In the present exploratory study, we evaluated the soluble compartment of BM and peripheral blood (PB) in 16 ALL pediatric patients by the means of the SOMAscan assay, a highly multiplexed, affinity proteomics platform able to measure 1305 proteins in as little as 65µl of sample using modified protein-binding single-stranded DNA aptamers called SOMAmers, revealed on a high dimension oligo-probe array. The raw hybridization data were normalized using hybridization controls and were log2-transformed. Transformed HybNormalized BM and PB plasma data were compared using GraphPad PRISM 8. Multiple comparisons were set with an FDR threshold of 0.01%.
Results
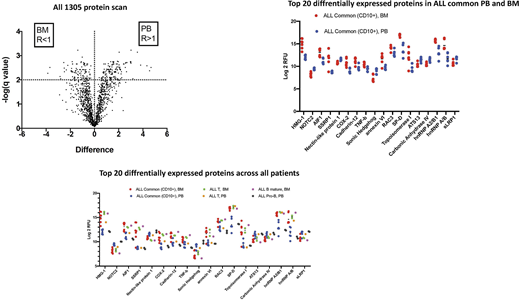
The comparative analysis between plasma samples from BM and PB showed that 228 proteins are differentially expressed in BM and PB plasma irrespective of the nature of disease (fig1). A more detailed mix-effect analysis of protein expression in the different patient categories and compartment defined in Table 1 revealed that out of these 228 proteins, 92 showed a significant differential expression when performing a mixed-effect analysis between the different groups. Among the top 20 proteins differentially expressed between BM and PB, 18 show differential expression between patient groups. The patient group effect is dominated by the differences between BM and PB plasma in ALL Common patients , which account for 17 of the observed differences in protein levels. In spite of the limited sample size, the analysis revealed a difference in NOTC2 level between ALL Common PB and ALL Pro-B PB.
Conclusion
In our study we applied a quantitative large-scale proteomic analysis on BM and PB plasma in children suffering from ALL, identifying differential protein expression between the two compartments.
In the last few years, SomaScan has emerged as a very attractive method due to its high throughput capacity, faster discovery mode compared to other proteomic platforms and small sample size required, which make it ideal for the identification of novel biomarkers. In spite of the limiting samples sizes, some differences in protein expression were revealed. We are taking this analysis further by analyzing more patients in different groups and including normal donor samples.
Table 1
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal